A decision tree classifier.
More...
#include <decision_tree.h>
|
| | DecisionTreeClassifier (std::vector< std::vector< std::string >> const &classes, std::vector< std::string > const &features, std::string const &class_balance="balanced", TreeDepthIdx_t max_depth=0, FeaturesIdx_t max_features=0, unsigned long max_thresholds=0, std::string const &missing_values="None", long random_state_seed=0) |
| | Create and initialize a new decision tree classifier. More...
|
| |
| void | fit (std::vector< Features_t > &X, std::vector< Classes_t > &y) |
| | Build a decision tree classifier from the training data. More...
|
| |
| void | predict_proba (Features_t *X, SamplesIdx_t n_samples, double *y_prob) |
| | Predict classes probabilities for the test data. More...
|
| |
| void | predict (Features_t *X, SamplesIdx_t n_samples, Classes_t *y) |
| | Predict classes for the test data. More...
|
| |
| double | score (Features_t *X, Classes_t *y, SamplesIdx_t n_samples) |
| | Calculate score for the test data. More...
|
| |
| void | calculate_feature_importances (double *importances) |
| | Calculate feature importances from the decision tree. More...
|
| |
| void | export_graphviz (std::string const &filename, bool rotate=false) |
| | Export of a decision tree in GraphViz dot format. More...
|
| |
| std::string | export_text () |
| | Export of a decision tree in a simple text format. More...
|
| |
| void | export_serialize (std::string const &filename) |
| | Export of a decision tree classifier in binary serialized format. More...
|
| |
| void | serialize (std::ofstream &fout) |
| | Serialize. More...
|
| |
A decision tree classifier.
◆ DecisionTreeClassifier()
| koho::DecisionTreeClassifier::DecisionTreeClassifier |
( |
std::vector< std::vector< std::string >> const & |
classes, |
|
|
std::vector< std::string > const & |
features, |
|
|
std::string const & |
class_balance = "balanced", |
|
|
TreeDepthIdx_t |
max_depth = 0, |
|
|
FeaturesIdx_t |
max_features = 0, |
|
|
unsigned long |
max_thresholds = 0, |
|
|
std::string const & |
missing_values = "None", |
|
|
long |
random_state_seed = 0 |
|
) |
| |
Create and initialize a new decision tree classifier.
- Parameters
-
| [in] | classes | Class labels for each output. |
| [in] | features | Feature names. |
| [in] | class_balance | Weighting of the classes.
string "balanced" or "None", (default="balanced")
If "balanced", then the values of y are used to automatically adjust class weights inversely proportional to class frequencies in the input data.
If "None", all classes are supposed to have weight one. |
| [in] | max_depth | The maximum depth of the tree.
The depth of the tree is expanded until the specified maximum depth of the tree is reached or all leaves are pure or no further impurity improvement can be achieved.
integer (default=3)
If 0 the maximum depth of the tree is set to max long (2^31-1). |
| [in] | max_features | Number of random features to consider when looking for the best split at each node, between 1 and n_features.
Note: the search for a split does not stop until at least one valid partition of the node samples is found up to the point that all features have been considered, even if it requires to effectively inspect more than max_features features.
integer (default=0)
If 0 the number of random features = number of features.
Note: only to be used by Decision Forest |
| [in] | max_thresholds | Number of random thresholds to consider when looking for the best split at each node.
integer (default=0)
If 0, then all thresholds, based on the mid-point of the node samples, are considered.
If 1, then consider 1 random threshold, based on the Extreme Randomized Tree formulation.
Note: only to be used by Decision Forest |
| [in] | missing_values | Handling of missing values.
string "NMAR" or "None", (default="None")
If "NMAR" (Not Missing At Random), then during training: the split criterion considers missing values as another category and samples with missing values are passed to either the left or the right child depending on which option provides the best split, and then during testing: if the split criterion includes missing values, a missing value is dealt with accordingly (passed to left or right child), or if the split criterion does not include missing values, a missing value at a split criterion is dealt with by combining the results from both children proportionally to the number of samples that are passed to the children during training.
If "None", an error is raised if one of the features has a missing value.
An option is to use imputation (fill-in) of missing values prior to using the decision tree classifier. |
| [in] | random_state_seed | Seed used by the random number generator.
integer (default=0)
If -1, then the random number generator is seeded with the current system time.
Note: only to be used by Decision Forest |
"Decision Tree": max_features=n_features, max_thresholds=0.
The following configurations should only be used for "decision forests":
"Random Tree": max_features<n_features, max_thresholds=0.
"Extreme Randomized Trees (ET)": max_features=n_features, max_thresholds=1.
"Totally Randomized Trees": max_features=1, max_thresholds=1, very similar to "Perfect Random Trees (PERT)".
◆ calculate_feature_importances()
| void koho::DecisionTreeClassifier::calculate_feature_importances |
( |
double * |
importances | ) |
|
Calculate feature importances from the decision tree.
- Parameters
-
| [in,out] | importances | Feature importances corresponding to all features [n_features].
The higher, the more important the feature. The importance of a feature is computed as the (normalized) total reduction of the criterion brought by that feature. |
◆ deserialize()
◆ export_graphviz()
| void koho::DecisionTreeClassifier::export_graphviz |
( |
std::string const & |
filename, |
|
|
bool |
rotate = false |
|
) |
| |
Export of a decision tree in GraphViz dot format.
- Parameters
-
| [in] | filename | Filename of GraphViz dot file, extension .gv added. |
| [in] | rotate | Rotate display of decision tree.
boolean (default=false)
If false, then orient tree top-down.
If true, then orient tree left-to-right.
Ubuntu:
sudo apt-get install graphviz
sudo apt-get install xdot
view
$: xdot filename.gv
create pdf, png
$: dot -Tpdf filename.gv -o filename.pdf
$: dot -Tpng filename.gv -o filename.png
Windows:
Install graphviz-2.38.msi from http://www.graphviz.org/Download_windows.php
START> "Advanced System Settings"
Click "Environmental Variables ..."
Click "Browse..." Select "C:/ProgramFiles(x86)/Graphviz2.38/bin"
view
START> gvedit |
◆ export_serialize()
| void koho::DecisionTreeClassifier::export_serialize |
( |
std::string const & |
filename | ) |
|
Export of a decision tree classifier in binary serialized format.
- Parameters
-
| [in] | filename | Filename of binary serialized file, extension .dt added. |
◆ export_text()
| string koho::DecisionTreeClassifier::export_text |
( |
| ) |
|
Export of a decision tree in a simple text format.
◆ fit()
| void koho::DecisionTreeClassifier::fit |
( |
std::vector< Features_t > & |
X, |
|
|
std::vector< Classes_t > & |
y |
|
) |
| |
Build a decision tree classifier from the training data.
- Parameters
-
| [in] | X | Training input samples [n_samples x n_features]. |
| [in] | y | Target class labels corresponding to the training input samples [n_samples x n_outputs]. |
◆ import_deserialize()
Import of a decision tree classifier in binary serialized format.
- Parameters
-
| [in] | filename | Filename of binary serialized file. |
◆ predict()
Predict classes for the test data.
- Parameters
-
| [in] | X | Test input samples [n_samples x n_features]. |
| [in] | n_samples | Number of samples in the test data. |
| [in,out] | y | Predicted classes for the test input samples [n_samples].
Using 1d array addressing for X and y to support efficient Cython bindings to Python using memory views. |
◆ predict_proba()
Predict classes probabilities for the test data.
- Parameters
-
| [in] | X | Test input samples [n_samples x n_features]. |
| [in] | n_samples | Number of samples in the test data. |
| [in,out] | y_prob | Class probabilities corresponding to the test input samples [n_samples x n_classes x n_classes_max]. We use n_classes_max to create a nice 3D array to hold the predicted values x samples x classes as the number of classes can be different for different outputs.
Using 1d array addressing for X and y_prob to support efficient Cython bindings to Python using memory views. |
◆ score()
Calculate score for the test data.
- Parameters
-
| [in] | X | Test input samples [n_samples x n_features]. |
| [in] | y | True classes for the test input samples [n_samples]. |
| [in] | n_samples | Number of samples in the test data. |
- Returns
- Score.
Using 1d array addressing for X and y to support efficient Cython bindings to Python using memory views.
◆ serialize()
| void koho::DecisionTreeClassifier::serialize |
( |
std::ofstream & |
fout | ) |
|
◆ class_balance
| std::string koho::DecisionTreeClassifier::class_balance |
|
protected |
◆ classes
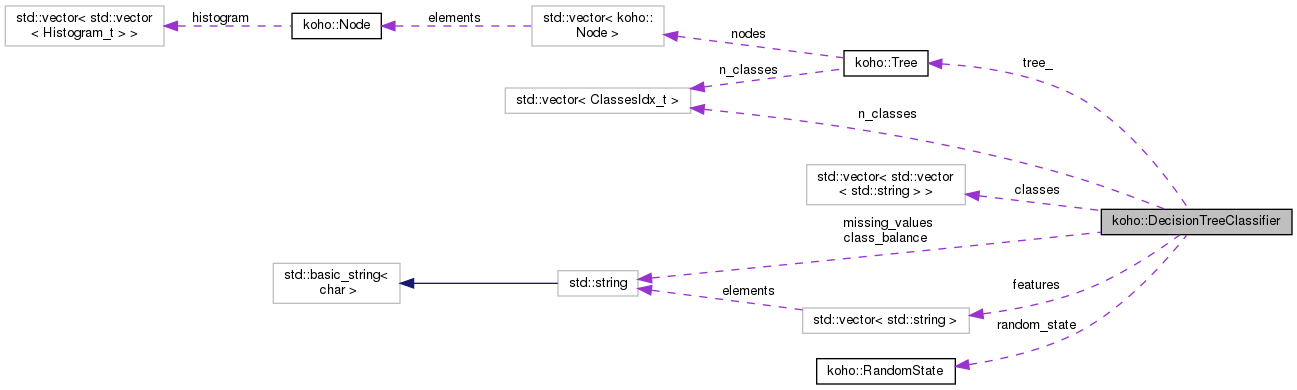
| std::vector<std::vector<std::string> > koho::DecisionTreeClassifier::classes |
|
protected |
◆ features
| std::vector<std::string> koho::DecisionTreeClassifier::features |
|
protected |
◆ max_depth
◆ max_features
◆ max_thresholds
| unsigned long koho::DecisionTreeClassifier::max_thresholds |
|
protected |
◆ missing_values
| std::string koho::DecisionTreeClassifier::missing_values |
|
protected |
◆ n_classes
| std::vector<ClassesIdx_t> koho::DecisionTreeClassifier::n_classes |
|
protected |
◆ n_classes_max
◆ n_features
◆ n_outputs
◆ random_state
◆ tree_
| Tree koho::DecisionTreeClassifier::tree_ |
|
protected |
The documentation for this class was generated from the following files:
 1.8.13
1.8.13